Double stranded DNA is a dynamic structure and should never be considered as a static entity. The two strands are held together by non-covalent interactions (hydrogen bonding and base stacking). The energy of these interactions is such that the helix can come apart quite easily at physiological temperatures. If this were not so, gene expression would not be possible at the temperature of living systems.
DNA can be heated and, at a certain temperature, the two strands will come apart. We say that the DNA helix has melted or denatured.
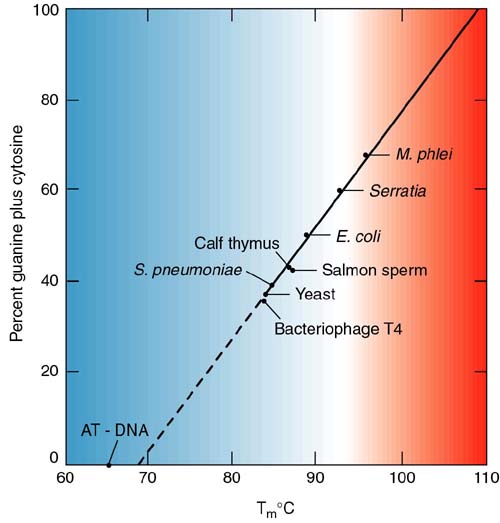
This transition can be followed by the increase in the absorption of ultraviolet light by the molecule as it goes from helix to random coil (the denatured form). This is called hyperchromicity:

The increase in UV absorbance at 260 nm for the denatured (coil) is used to follow the transition from helix to coil.
Here is an example from your book, using Streptococcus pneumoniae DNA (Figure 2.17):

The temperature at the midpoint of the transition is called the melting temperature or Tm.
As I'm sure you remember, AT base pairs have two hydrogen bonds and GC pairs have three. Therefore, there is more energy in the GC bond. Consequently, the higher the GC content of a molecule, the higher the melting temperature (more energy is necessary to make the helix-coil transition).
Which of the following two DNA molecules would have a lower melting temperature (Tm) than the other?
|
#1 |
#2 |
|
|
|
ANSWER:
Once the DNA strands have been melted away from each other, they can be allowed to come back together, to anneal or renature, by lowering the temperature (given the correct ionic strength of the solution). This process is also a kinetic one.
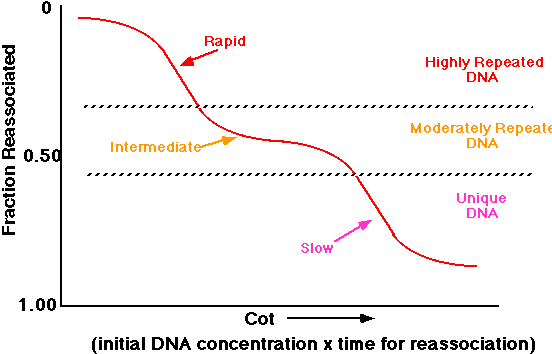
Without going into the mathematics in detail, the reaction is a second order rate, dependent upon the concentration of DNA. This can be expressed graphically as the Cot (we say "cot" just like the foldaway bed). Co refers to the initial DNA concentration and t the time of renaturation. If you plot the amount of the DNA that has renatured (reassociated) against the Cot, you get curves such as shown in Figure 2.20:

The inflection point of the curves is called the 1/2 Cot. Notice that the 1/2 Cot gets larger for more complex (larger) nucleic acids. The curve for the simplest molecule, poly(U):poly(A) [a synthetic structure] gives a value of 1 nucleotide pair. This is because the sequences match along their entire length and are effectively only one nucleotide long. All of these curves represent molecules where the sequence of the DNA is unique. That is, in the population there is no repeat of any sequence.
Eukaryotic DNA is much different than this simple picture. On the far right of the above figure you see calf DNA, the non-repetitive fraction. The total DNA has different populations of sequences. The Cot curve of total calf DNA is shown in Figure 2.21
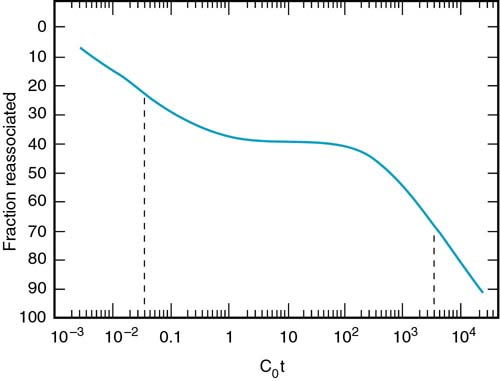
This can be interpreted using the following analysis. Suppose that we look at this population of DNA. We can see three different classes of sequences.
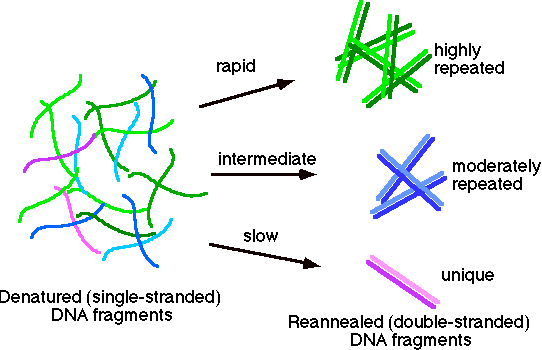
The highly repeated sequences (many copies per genome) will renature rapidly, since kinetically any strands can find a match much more quickly. The moderately repeated fraction will have an intermediate time of renaturation. Finally the unique sequence (e.g., calf non-repetitive fraction) will have the slowest time of renaturation.
The Cot curve for this population might be interpreted like this: